Antimicrobial Peptides (AMPs) or Host Defense Peptides (HDPs) are small organic molecules (5 to 50 amino acid residues) produced by organisms or synthetically designed. In nature, this is an essential component of any innate immune system. They are prospective candidates for novel therapeutic agents[1],[2],[3],[4],[5],[6] and are in use in conventional therapy as:
- An antibiotic with no antimicrobial resistance (AMP) against bacterium, protozoa, fungus, and yeasts, or other pathogenic micro-organisms;
- Anti-biofilm agent (ABP) for biotechnological and medical devices disinfection;
- Anti-viral (AVP) and viral infection prevent (HIV as well!) medicines;
- Immunomodulators and vaccines (HDP) for indirect activities, including use in autoimmune diseases;
- Cell-Penetration agent (CPP) to destroy pathogen and cancer cells and as a trans cell-membrane transport for drugs;
- Anti-inflammatory, pain modulating, neuroprotection in neurodegenerative diseases, etc.
 As a rule, peptides exhibit several of these properties at once.
As a rule, peptides exhibit several of these properties at once.
A large number of reviews and articles are devoted to the description of AMPs, the mechanisms of their action and medical perspectives. The search for “AMPs” on the PubMed portal demonstrates a rapid increase in interest in Antibacterial peptides.
For copyright reasons we try to reference general and newest but quote open access materials only.
After AMPs bind to target cell membranes through electrostatic interactions two general mechanisms [1] of action starts:
- To disrupt the cell membrane;
- To enter and inhibit intracellular functions;
- To modulate host immunity.
First founded and the most studied mechanism is direct membrane destruction. Non-membrane disruptive mechanisms are characterized by pathogen targeting, leading to their destruction, without membrane disruption.
[1] E. Neil G. Marsh, Benjamin C. Buera, Ayyalusamy Ramamoorthy. Fluorine—a new element in the design of membrane-active peptides. Mol. BioSyst (2009). DOI: 10.1039/B909864J. AMs biological functions infographics.
Cell surface targets:
- Interaction of peptides with different glycosaminoglycan (e.g., HS) present on the cell surface competing with the virus for cellular binding sites.
- Blocking of viral entry into the cell by binding the peptide to viral CXCR4co-receptor required for its entry.
- Suppression of cell fusion by interfering with the activity of ATPase protein.
- Suppression viral gene expression.
- Inhibition of peptide chain elongation by inactivating the ribosome.
- Activation of an immune modulatory pathway by induction of NK and IFN.
- Binding of peptides to viral proteins causing inhibition of adsorption/virus-cell fusion.
Mechanisms of actions of cationic antiviral peptides [1]
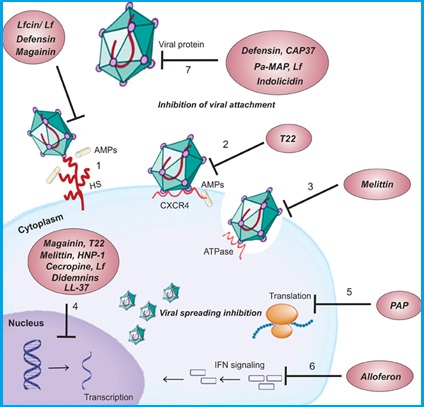
Cell surface targets: (1) Interaction of peptides with different glycosaminoglycan (e.g., HS) present on the cell surface competing with the virus for cellular binding sites. (2) Blocking of viral entry into the cell by binding the peptide to viral CXCR4co-receptor required for its entry. (3) Suppression of cell fusion by interfering with the activity of ATPase protein. Intracellular targets: (4) Suppression viral gene expression. (5) Inhibition of peptide chain elongation by inactivating the ribosome. (6) Activation of an immune modulatory pathway by induction of NK and IFN. Viral protein targets: (7) Binding of peptides to viral proteins causing inhibition of adsorption/virus-cell fusion.
[1] Mulder Kelly, Lima Loiane Alves, Miranda Vivian, Dias Simoni, Franco Octavio. Current scenario of peptide-based drugs: the key roles of cationic antitumor and antiviral peptides. Frontiers in Microbiology (2013) DOI: 10.3389/fmicb.2013.00321
The model for AMPs membrane permeation [1],[2],[3]
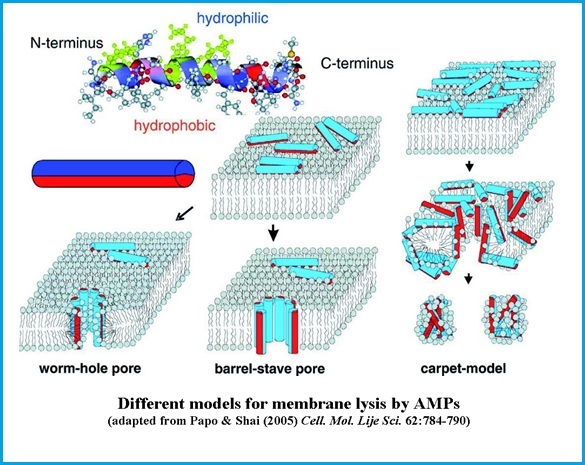
Sketch of different models describing the functional mechanisms of linear antimicrobial peptides interacting with lipid bilayers. (Top, left) Many AMPs have the potential to fold into amphipathic a-helices with hydrophilic and hydrophobic sides. This confirmation is here schematically represented as an amphiphilic cylinder, with hydrophobic (red) and hydrophilic (blue) halves. (Center) Antimicrobial peptides bind to the surface of the membrane with the hydrophobic side groups anchored in the hydrophobic lipidic core of the bilayer, leading to different outcomes. (Bottom, left) The wormhole (or toroidal) pore model as proposed for magainin (Bottom, center) Barrel-stave pore model, as proposed for alamethicin, a peptide antibiotic produced by the soil fungus Trichoderma Viridis. (Bottom, right) Carpet model: antimicrobial peptides crowd on the membranes surface and lead subsequently to micellization.
The antimicrobial peptide database (APD) http://aps.unmc.edu/AP/main.php is the most extensive online database. It contains nearly 3000 AMPs separated from organisms of
AMPs are separated from organisms of six kingdoms:
- 303 bacteriocins/peptide from bacteria,
- 4 from archaea,
- 8 from protists,
- 13 from fungi,
- 342 from plants
- 2181 from animals


with the following activities:
- Antibacterial peptides;
- Antibiofilm peptides;
- Antiviral (and Anti-HIV) peptides;
- Antifungal peptides;
- Antiparasitic peptides;
- Antimalarial peptides;
- Anti-protist peptides;
- Anticancer peptides;
- Antioxidant peptides;
- Chemotactic peptides;
- Insecticidal peptides;
- Protease inhibitors;
- Spermicidal peptides;
- Surface immobilized peptides;
- Wound healing peptides
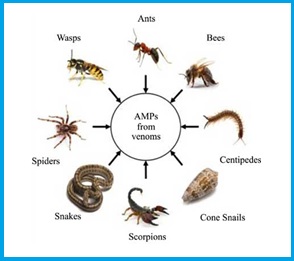 The peptide components of venoms [1],[2] from Hymenoptera (bees, wasps, and ants) are spread over the molar mass range of 1400 to 7000 Da and together comprise up to 70% of the weight of freeze-dried venoms.
The peptide components of venoms [1],[2] from Hymenoptera (bees, wasps, and ants) are spread over the molar mass range of 1400 to 7000 Da and together comprise up to 70% of the weight of freeze-dried venoms.
Most of these toxins are linear polycationic amphipatic peptides with a high content of α-helices in their secondary structures.
These peptides generally account for cell lysis, hemolysis, antibiosis, and sometimes promote the delivery of cellular activators/mediators through interaction with the G-protein receptor, and perhaps some of them are even immunogenic components.
Current Topics in Medicinal Chemistry. DOI: 10.2174/1568026616666160930151242
The AMPs field is growing rapidly, coherently with the whole antibiotics market [1] in response to the demand for novel antimicrobial agents for drug-resistant pathogens. The rise in research activities [2] and development pipeline of peptide drugs is projected to bring growth. The Global Peptide Therapeutics Market research [3] (Published Date: 2017-04-26) project the opportunity in this market to expand at a healthy CAGR [4] of 9.10% and reach a value of US$46.6 bn. by the end of 2024.
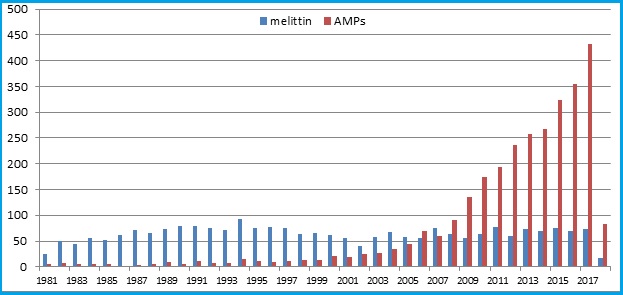
The search for “AMPs” and “Melittin” on the PubMed portal demonstrates a stable interest in the study of melittin and a rapid increase in interest in Antibacterial peptides.
Search results by year [5]
Cancer Treatment to Surface as Key Application Area of Peptide Therapeutics
Based on the application, the global peptide therapeutics market is classified into cancer treatment, drugs for metabolic disorders, cardiovascular disease (CVD) therapeutics, respiratory disorder therapeutics, gastrointestinal tract (GIT) therapeutics, anti-infective drugs, dermatology therapeutics, central nervous systems (CNS) therapeutics, and renal therapeutics.
With peptides evolving as a potential therapeutic agent for the treatment of cancer, the cancer segment emerged as the key contributor to this market in 2015. The rising mortality rate, caused by cancer, has led to an increased focus of players towards the development of new drugs and products to combat the situation, which is influencing the market substantially. The technological advancements and extensive research and development activities are likely to fuel the uptake of peptide therapeutics for the treatment of cancer over the next few years.
Peptide therapeutics mainly follow two routes of administration: Oral and parenteral. The parenteral segment led the global market for peptide therapeutics in 2015 due to the easy availability of peptide drugs for the parenteral route of administration compared to other routes. However, the oral administration of these drugs is likely to increase significantly over the next few years.
Based on the application, the global peptide therapeutics market is classified into cancer treatment, drugs for metabolic disorders, cardiovascular disease (CVD) therapeutics, respiratory disorder therapeutics, gastrointestinal tract (GIT) therapeutics, anti-infective drugs, dermatology therapeutics, central nervous systems (CNS) therapeutics, and renal therapeutics.
With peptides evolving as a potential therapeutic agent for the treatment of cancer, the cancer segment emerged as the key contributor to this market in 2015. The rising mortality rate, caused by cancer, has led to an increased focus of players towards the development of new drugs and products to combat the situation, which is influencing the market substantially. The technological advancements and extensive research and development activities are likely to fuel the uptake of peptide therapeutics for the treatment of cancer over the next few years.
Peptide therapeutics mainly follows two routes of administration: Oral and parenteral. The parenteral segment led the global market for peptide therapeutics in 2015 due to the easy availability of peptide drugs for the parenteral route of administration compared to other routes. However, the oral administration of these drugs is likely to increase significantly over the next few years.
Continued Dominance of North America in Global Peptide Therapeutics Market
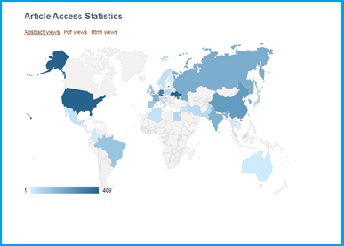 Geographically, the worldwide market for peptide therapeutics has been segmented into North America, Europe, Asia Pacific, the Middle East and Africa, and Latin America. North America led the global market with a share of 38.8% in 2015 and is expected to maintain its dominance over the next few years. The advancements in the healthcare infrastructure and the advent of peptide therapeutics as a highly potent drug used in oncology and hormonal therapies are likely to boost the growth of the North America peptide therapeutics market in the near future.
Geographically, the worldwide market for peptide therapeutics has been segmented into North America, Europe, Asia Pacific, the Middle East and Africa, and Latin America. North America led the global market with a share of 38.8% in 2015 and is expected to maintain its dominance over the next few years. The advancements in the healthcare infrastructure and the advent of peptide therapeutics as a highly potent drug used in oncology and hormonal therapies are likely to boost the growth of the North America peptide therapeutics market in the near future.
Apart from this, the presence of well-established players in the U.S. is also projected to support this regional market over the forthcoming years. Europe, which stood second in 2015, is also anticipated to witness healthy growth in the years to come.
The global peptide therapeutics market is extremely consolidated in nature. At the forefront of this market are Teva Pharmaceutical Industries Ltd., Takeda Pharmaceutical Co. Ltd., Sanofi, Pfizer Inc., Novartis AG, GlaxoSmithKline Plc, F. Hoffmann-La Roche Ltd., Eli Lilly and Co., Bachem Holding AG, and Amgen Inc.
A geographical distribution of AMPs field interest illustration is the access map[6] for a review article Sok Cheon Pak. An Introduction to the Toxins Special Issue on “Bee and Wasp Venoms: Biological Characteristics and Therapeutic Application”. Toxins (2016) DOI: 10.3390/toxins8110315. The data may have bias regards to the language of publications.
In order to obtain a broad market approval honeybee venom and it’s derivative melittin needs standardization that would guarantee its quality and repeatable pharmacological activity.
Main procedures of analysis are aimed on determine purity, bio-chemical composition and bio-medical properties of the product.
- High-Performance Liquid Chromatography (HPLC) [1] methods identify the peptides components purity and it’s proportion in the product.
- Mass spectrometry (MS) [2] techniques identify precisely the particular chemical structure of the components and the product.
- Electrophoresis [3] (SDS-PAGE) [4] effectively determine an existence of melittin and other components in the product.
- Pharm-tests [5],[6]: Hemolytic & Antimicrobial Activity, Histamine Releasing Activity, HYA Enzyme Assay, etc. show the bio-medical activity of the melittin in the product.
- Biophysical methods [7] are experimental and simulation techniques to study the structure, properties, dynamics or function of biomolecules at an atomic or molecular level.
There are numerous publications concern analysis and standardization [8],[9] of the Bee Venom and its derivatives, first of all melittin. This section also describes some specific technologies necessary for the product storage and perspective of its application.
High Performance Liquid Chromatography (HPLC) method is fruitful for separate [1], identify & quantify components dissolved in a liquid solvent. The quality of the analytical resolution depends as far as the chromatogram allows to separate and to identify different peaks. The retention time (depends on equipment) – peak itself, identify the component. The Area% – the area of each peak in the chromatogram as a percentage of the total area of all peaks provides a suitable approximation of the relative amounts of components in the sample [2]. Usually Area% is calculated by the chromatograph software.
The typical chromatogram of the melittin samples [3]
The standard (Sigma-Aldrich) synthetic melittin sample
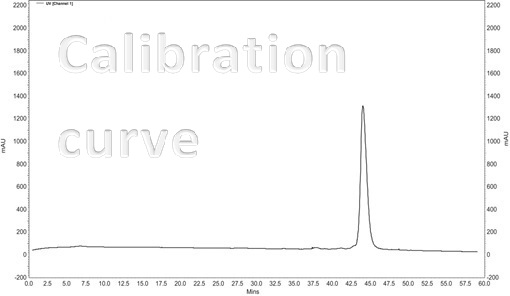
The curve of the purified sample of natural origin
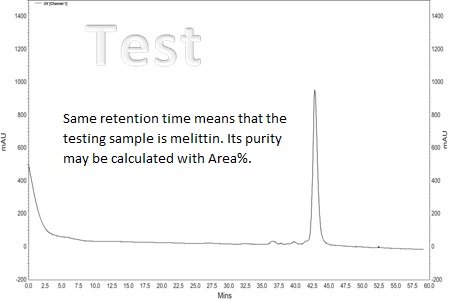
There are lot of clear materials about HPLC [4] and how to read Chromotogram[5]
Mass Spectrometry analysis is often used to identify a specific peak in the chromatogram after the peptide mixture is HPLC fractionated.
The typical Mass Specter of the melittin sample [1]
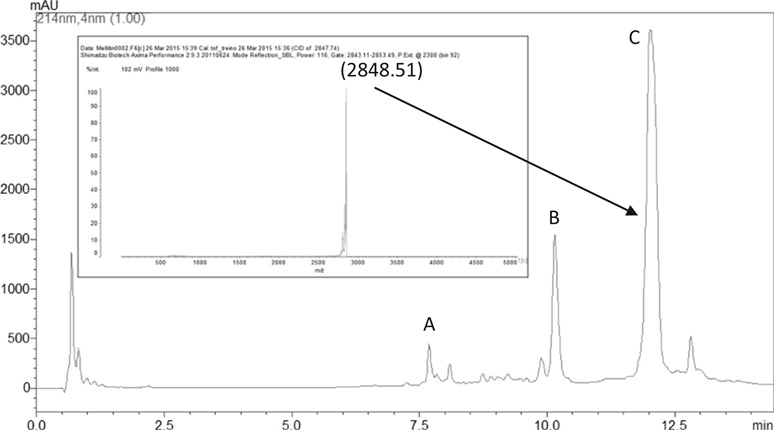
The HPLC of the whole venom, identifying the venom major peaks, namely: apamin (A), phospholipase A2 (B) and melittin (C). Peak C was analyzed by mass spectrometry (inset) and demonstrated to contain pure melittin.
[1] Andreia Vieira Pereira, Gustavo de Barros, Erika Gracielle Pinto, Andre Gustavo Tempone, Ricardo de Oliveira Orsi, Lucilene Delazari dos Santos, Sueli Calvi, Rui Seabra FerreiraJr, Daniel Carvalho Pimenta, Benedito Barraviera. Melittin induces in vitro death of Leishmania (Leishmania) infantum by triggering the cellular innate immune response Journal of Venomous Animals and Toxins including Tropical Diseases (2016) DOI: 10.1186/s40409-016-0055-x
Electrophoresis is an affordable and cheap method for analyzing complex protein mixtures. This makes it possible to effectively determine the purity of proteins in the HPLC fraction with a minimum amount of the sample.[1]
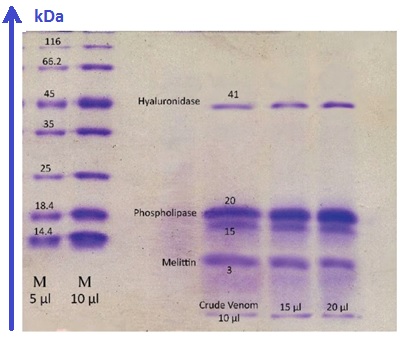
The typical Electrophoresis (SDS-PAGE [2]) of the crude venom and melittin sample [3]
Apis mellifera crude venom.
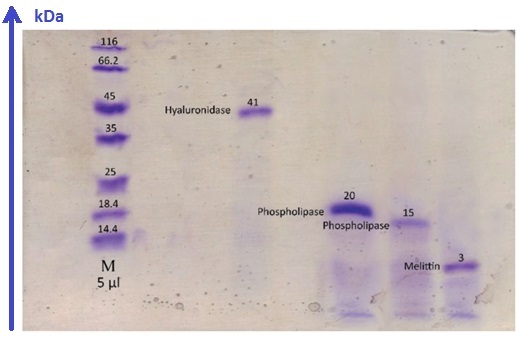
Apis mellifera HPLC fractions
The goal of AMPs isolating from biological substances is to prepare some biologically active chemicals with certain medical properties. However, during storage and chemical processing, the properties of peptides may vary or disappears. Therefore it is important to check the biological properties at different stages of the process, from testing the raw material to testing the properties of the final products. Methods for testing the biological activity of drugs have been known since ancient times. Different BiologicalTests/BioAssay and results for melittin are mentioned at PubMed [1].
The most common pharmacological test for melittin is Hemolytic & Antimicrobial Activities assays. In general, the hemolytic activity used to evaluate of potential bacteria or drugs and unknown substances in the destruction of red blood cells. In addition, one of the trials evaluating the safety of drug dose is hemolytic activity assay. Examples of Bee Venom and melittin pharm-test protocols may be found in numerous articles.[2],[3],[4] Antimicrobial activity assay examples.[5],[6] Hemolytic activity assay examples.[7],[8] Histamine-releasing activity assay example [9]
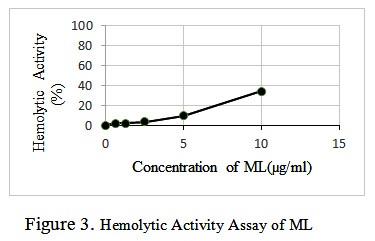 The hemolytic activity of bee venom and ML was assayed by a standard procedure[10]. In concise, fresh rabbit red blood cells (RBCs) that were collected in the presence of an anticoagulant (sodium citrate) were washed three times in PBS. ML (in selected amounts dissolved in water 0.1, 0.05, 0.001 µg/ml, respectively) were added to the suspension of red blood cells (2% v/v) in PBS to the final volume of 200 μl and incubated at 37 °C for 35 min. The samples were centrifuged for 10 min at 2000 rpm, and the release of hemoglobin was monitored by measuring the absorbance (Asampel) of the supernatant at 540 nm. RBCs in PBS (Ablank) and in 0.2% v/v Triton X-100 (ATriton) were used for negative and positive controls respectively. The percentage of hemolysis was calculated according to the equation, Percentage of hemolysis = [(Asampel – Ablank)/( ATriton – Ablank)] × 100.
The hemolytic activity of bee venom and ML was assayed by a standard procedure[10]. In concise, fresh rabbit red blood cells (RBCs) that were collected in the presence of an anticoagulant (sodium citrate) were washed three times in PBS. ML (in selected amounts dissolved in water 0.1, 0.05, 0.001 µg/ml, respectively) were added to the suspension of red blood cells (2% v/v) in PBS to the final volume of 200 μl and incubated at 37 °C for 35 min. The samples were centrifuged for 10 min at 2000 rpm, and the release of hemoglobin was monitored by measuring the absorbance (Asampel) of the supernatant at 540 nm. RBCs in PBS (Ablank) and in 0.2% v/v Triton X-100 (ATriton) were used for negative and positive controls respectively. The percentage of hemolysis was calculated according to the equation, Percentage of hemolysis = [(Asampel – Ablank)/( ATriton – Ablank)] × 100.
Biophysical methods are exact sciences techniques to study the structure, properties, dynamics or function of biomolecules at an atomic or molecular level. They encompass a range of experimental and theoretical techniques including microscopy, spectroscopy, electrophysiology, single-molecule methods, Molecular Dynamic (MD) simulation and Molecular Docking modeling. Computational approaches of Proteome Bioinformatics [1] complement experimental techniques.
Membrane-active antimicrobial peptides as melittin are challenging to study experimentally, but relatively easy to investigate using molecular dynamics (MD) computer simulations and Molecular Docking modelling. For this reason, a large number of studies have been reported over recent years. Molecular Dynamic (MD) simulation example may be found.[2]
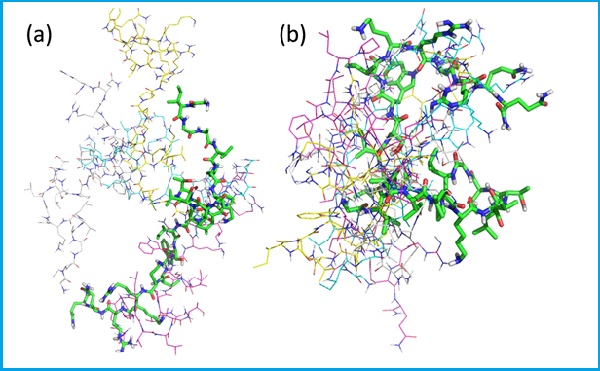
Molecular Docking modeling example [3].
Super-imposition of the five best docking poses of melittin with lecithin PC and PS67–b-PAA27 polymer:
Biophysical Techniques [4]
The characterization of molecular structure, the measurement of molecular properties, and the observation of molecular behavior presents an enormous challenge for biological scientists. A wide range of biophysical techniques has been developed to study molecules in crystals, in solution, in cells, and in organisms. These biophysical techniques provide information about the electronic structure, size, shape, dynamics, polarity, and modes of interaction of biological molecules. Some of the most exciting techniques provide images of cells, subcellular structures, and even individual molecules. It is now possible, for example, to directly observe the biological behavior and physical properties of the single protein or DNA molecules within a living cell and determine how the behavior of the single molecule influences the biological function of the organism.
Much biophysical research involves either the development of novel techniques to investigate the structure, properties, and biological functions of biomolecules or the application of these techniques to monitor how the structure and dynamics of biomolecules enable specific biological functions. Information about specific biophysical techniques is provided here.
Electrophysiology
- Detection of Secretion by Electrochemical Methods, Spencer Hochstetler, and R. Mark Wightman. Previously published in Biophysics Textbook Online (BTOL).
Hydrodynamics
The behavior of large biomolecules—proteins, carbohydrates, and nucleic acids—in solution is complex and directly related to molecular size, shape, and flexibility; the analysis of hydrodynamic behavior thus provides important information about the structure, dynamics, and interactions of biomacromolecules.
- Survey of Biomolecular Hydrodynamics, Victor Bloomfield. Previously published in Biophysics Textbook Online (BTOL).
Microscopy & Imaging
Perhaps the most accessible developments in biophysics have involved improvements in our ability to generate images of cellular and molecular structures with dimensions from microns to nanometers. It is now possible to “see” individual molecules or cellular structures using atomic force, electron, or confocal fluorescence microscopy.
- Molecular Expressions: Exploring the World of Optics and Microscopy, a delightful and extensive site appropriate for middle school to college/university levels containing a photo gallery of microscopic images, a detailed primer on the basics of optical microscopy, as well as historical and educational information on optical microscopy. Generated and maintained by the National High Field Magnet Laboratory at Florida State University.
Confocal Fluorescence Microscopy
- Nikon Microscopy U, an extensive and well-organized site with information on the fundamentals of confocal fluorescence microscopy and its applications to biological systems. Content provided by various experts and site maintained by Nikon USA.
Atomic Force Microscopy
- Atomic Force Microscopy: Measuring Intermolecular Interaction Force, a primer on the fundamentals and applications of AFM by David Baselt.
Magnetic Resonance Imaging
- Magnetic Resonance Imaging offers information on the biophysical foundations and medical applications of one of the most versatile of medical imaging tools; provided as part of the Breakthroughs in Science series of FASEB, the Federation of American Societies for Experimental Biology.
Modeling & Simulation
Visual and numerical modeling and simulation play important roles in the analysis and prediction of protein and nucleic acid sequence and 3D structure, in the calculation of molecular dynamics, and in the simulation of specific biochemical mechanisms, among other important tasks.
- Protein Structure Prediction From Primary Sequence, Linda Ellis and Kim-Hung Chow. Previously published in Biophysics Textbook Online (BTOL).
- Introduction to Continuum Electrostatics, with Molecular Applications, Mike Gilson. Previously published in Biophysics Textbook Online (BTOL).
- A Molecular Modeler’s Guide to Statistical Mechanics, Daniel A. Beard. Previously published in Biophysics Textbook Online (BTOL).
- Mathematical Models in Biophysics, Galina Riznichenko. Previously published in Biophysics Textbook Online (BTOL).
- Introduction to Macromolecular Simulation, Peter Steinbach. Previously published in Biophysics Textbook Online (BTOL).
- Berkeley Madonna is a fast, easy to use mathematical model building program that runs on both Windows and Mac OS. Using ordinary math notation, it provides numerical solutions to differential/difference equations together with a number of devices for analyzing results without any programming input from the user. Developed on the Berkeley campus under the sponsorship of NSF and NIH, it is currently used by academic and commercial institutions for both research and teaching. A visual tour of the program as well as a demo version can be downloaded from the website.
Single Molecule Techniques
Perhaps the most exciting development in biophysical technique in recent times involves the ability to manipulate single molecules and measure their properties and biological functions both in solution and within cells.
- Molecules in Action, Christopher Miller. Previously published in Biophysics Textbook Online (BTOL).
- Fluorescence Correlation Spectroscopy, Petra Schwille and Elke Haustein. Previously published in Biophysics Textbook Online (BTOL).
- Optical Tweezers: Measuring Piconewton Forces, Mark Williams. Previously published in Biophysics Textbook Online (BTOL).
Spectroscopy
The interaction of electromagnetic radiation, x-rays, ultraviolet, visible, and infra red light, and radio waves, with molecules provides a wealth of information about the structure, dynamics, and function of biomolecules and biological processes.
Fluorescence
-
Approaches To Teaching Fluorescence Spectroscopy, Catherine Royer. Previously published in Biophysical Journal and reprinted in Biophysics Textbook Online (BTOL).
-
Fluorescence Correlation Spectroscopy, Petra Schwille and Elke Haustein. Previously published in Biophysics Textbook Online (BTOL).
- Molecular Probes, Inc. is the major commercial source for fluorescent probes and diagnostic reagents. Their website provides spectroscopic data on a wide variety of molecular probes (with access to an extensive bibliography of biophysical applications), information about fluorescence techniques, as well as other resources for optical biophysics research.
Magnetic Resonance
- Fundamentals of NMR, Tom James. Previously published in Biophysics Textbook Online (BTOL).
- Pictorial Representation of Product-Operator Formalism: Non-Classical Vector Diagrams for Multidimensional NMR, David Donne and David Gorenstein. Previously published in Biophysics Textbook Online (BTOL).
- Electron-Nucleus Interactions and Its Biophysical Consequences, Ivano Bertini and Claudio Luchinat. Previously published in Biophysics Textbook Online (BTOL).
- NMR of Paramagnetic Proteins, Ivano Bertini and Claudio Luchinat. Previously published in Biophysics Textbook Online (BTOL).
- Fluorine NMR, J.T. Gerig. Previously published in Biophysics Textbook Online (BTOL).
- Teaching High-Resolution Nuclear Magnetic Resonance to Graduate Students in Biophysics, Laura Lerner and David Horita. Previously published in Biophysical Journal and reprinted in Biophysics Textbook Online (BTOL).
Thermodynamics & Related Topics
- Thermodynamics and Statistical Mechanics, Victor Bloomfield. Previously published in Biophysics Textbook Online (BTOL).
- Electrodynamic (van der Waals) Forces, Adrian Parsegian. Previously published in Biophysics Textbook Online (BTOL).
- Calculation of Biochemical Net Reactions and Pathways by Using Matrix Operations, Robert Alberty. Previously published in Biophysical Journal and reprinted in Biophysics Textbook Online (BTOL).
Some resource and data for computer simulation and modelling
Protein Data Bank (PDB) archive – information about the 3D structures of biological molecules (*.pdb files)
The DrugBank database is a unique bioinformatics and cheminformatics resource that combines detailed drug data with comprehensive drug target information.
PubChem, PubMed and other NCBI DB
ChemDB: a public database of small molecules and related chemoinformatics resources.
SDF (*.sdf files) is one of a family of chemical-data file formats developed by MDL; it is intended especially for structural information. “SDF” stands for structure-data file
OpenBabel converter (sdf -> pdb) (+H, 2D->3D)
Model Viewers
RasMol is a computer program written for molecular graphics visualization
RasTop is a molecular graphics program intended for the visualisation of proteins, nucleic acids and small molecules
ADT tools – AutoDockTools, is the ultimate GUI to view, set up, launch and analyze molecules in 3D, rotate & scale in real time.
Protein Structures Art – Shop for protein structure art and designs from the world’s greatest living artists.
Some Docking Modelling sofware
AutoDock VINA – molecular docking and virtual screening program
BLAST (Basic Local Alignment Search Tool) finds regions of similarity between biological sequences.
Some molecular dynamic MD simulation sofware
GROMACS is a versatile package to perform molecular dynamics, i.e. simulate the Newtonian equations of motion for systems with hundreds to millions of particles. (www.gromacs.org)
HIPPO is a software package for simulation and analysis of bio-molecules at an atomic level. (www.biowerkzeug.com)
Freeze-drying—technically known as lyophilisation, lyophilization, or cryodesiccation—is a dehydration process typically used to preserve a perishable material or make the material more convenient for transport. Freeze-drying works by freezing the material and then reducing the surrounding pressure to allow the frozen water in the material to sublime directly from the solid phase to the gas phase.
Freeze Dry Process [1]
Freeze drying is an important process in sample preparation and for the preservation and storage of biologicals, pharmaceuticals and foods. Of the various methods of dehydration, freeze drying (lyophilization) is especially suited for substances that are heat sensitive. Other than food processing (e.g., coffee, whole dinners), freeze drying has been extensively used in the development of pharmaceuticals (e.g., antibiotics) and preservation of biologicals (e.g., proteins, plasma, viruses and cell lines). The nondestructive nature of this process has been demonstrated by the retention of viability in freeze dried viruses and microorganisms.
Freeze drying is a process whereby water or other solvent is removed from frozen material by converting the frozen water directly into vapor without the intermediate formation of liquid water. The basis for this sublimation process involves the absorption of heat by the frozen sample in order to vaporize the ice; the use of a vacuum pump to enhance the removal of water vapor from the surface of the sample; the transfer of water vapor to a collector; and the removal of heat by the collector in order to condense the water vapor. In essence, the freeze dry process is a balance between the heat absorbed by the sample to vaporize the ice and the heat removed from the collector to convert the water vapor into ice.
[1] Ó’Fágáin C., Colliton K. Storage and Lyophilization of Pure Proteins. Springer Protocols (2017) DOI: 10.1007/978-1-4939-6412-3_9
The melittin therapeutic potential has not been fully achieved in clinic due to their off-target toxicity, rapid degradation and clearance in vivo. So the goal to incorporate melittin into synthetic polymer nanoparticles (NPs) with intrinsic affinity for target biomacromolecules hold great promise in the development of novel tools for biological and biomedical research.[1] ,[2]
In 2013 there were many publications about a breakthrough in this area[3], but real working medical technologies have not yet appeared on the market.
- S. National Library of Medicine® (NLM), The National Center for Biotechnology Information (NCBI) including:
- Springer Mainly scientific protocols “Methods in Molecular Biology”™ book series, including: Proteome Bioinformatics, Protein Chromatography, Computational Protein Design, etc.
- Antimicrobial Peptide Database (APD3)
- The Protein Data Bank (PDB) 3D structures of large biological molecules
- Hymenoptera Genome Database (HGD) bees, wasps, ants genomes, proteomes, etc.
- Web of Science online subscription-based scientific citation indexing service
- Google Scholar search engine for scientific information
- Scopus abstract and citation database of peer-reviewed research literature
- Cross Ref publications DOI index (Digital Object Identifier)
- CAS (Chemical Abstracts Service) – CAS RN index, including CommonChemistry open web resource that contains CAS Registry Numbers
- Hindawi peer-reviewed, fully Open Access journals (Evidence-Based Complementary and Alternative Medicine)
- MolBase integrated platforms for chemical e-commerce
- American Peptide Society (APEPS) http://www.americanpeptidesociety.org/
- Japanese Peptide Society https://www.peptide-soc.jp/en/
- The Protein Society (PS) http://proteinsociety.org
- Federation of American Societies for Experimental Biology (FASEB) http://org/home.aspx
- The Biophysical Society http://www.biophysics.org/
- American Society for Biochemistry and Molecular Biology http://www.asbmb.org/
- The International Biotherapy Society (IBS) http://biotherapysociety.org/

- Springer Protocols. Methods in Molecular Biology™ book series. Methods and Protocols.
- Paul Robert Hansen (Ed.) Antimicrobial Peptides (2017). DOI:1007/978-1-4939-6737-7
- Biji T. Kurien, R. Hal Scofield (Ed.) Protein Electrophoresis (2012). DOI:1007/978-1-61779-821-4.
- Andrea Giuliani, Andrea C. Rinaldi (Ed.) Antimicrobial Peptides (2010). DOI:1007/978-1-60761-594-1.
- Gregg B. Fields (Ed.) Peptide Characterization and Application Protocols (2007) DOI:1007/978-1-59745-430-8.
- Colin T. Mant, Yuxin Chen, Zhe Yan, Traian V. Popa, James M. Kovacs, Janine B. Mills et al. HPLC Analysis and Purification of Peptides. Pages 3-55
- Marie-Isabel Aguilar (Ed.) HPLC of Peptides and Proteins (2004) DOI:1385/1592597424.
- Parachin Nadia S., Franco Octavio L. New edge of antibiotic development: antimicrobial peptides and corresponding resistance. Research Topic. Frontier in Microbiology. (2014)
DOI: 3389/fmicb.2014.00147 pdf: frontiersin.org/research-topics - Amit Kessel, Nir Ben-Tal. Introduction to Proteins: Structure, Function, and Motion.
- Eric Martz. Introduction to proteins—structure, function, and motion.DOI: 10.1002/bmb.20603
- V. Finkelstein and O. B. Ptitsyn. Protein Physics. DOI: 10.1002/cbf.1064
- Handbook of Biologically Active Peptides (Second Edition), edited by Abba J. Kastin, Academic Press, Boston, 2013, ISBN 9780123850959, https://doi.org/10.1016/B978-0-12-385095-9.00077-4.
- PLOS One peer-reviewed resource